Lab homework assignment:
Express GFP in cell free system.
The kit
- enzyme mix at 1.33x (Please keep the enzyme mix frozen at all times - in -80 freezer or supplementing the dry ice until you're ready for experiment)
- plasmid P70-deGFP from Vincent Noireaux, same as used here: https://pubs.acs.org/doi/abs/10.1021/sb200016s
Experiment setup
1. Thaw the enzyme mix on ice (or in the fridge) immediately before use.
2. For 10uL reaction: use 7.5uL of enzyme mix, x*uL DNA vector to the final concentration 5nM, add water to the final reaction volume.
*We will know what's the stock c of DNA vector soon
3. Mix by gently pipetting up and down several times. Do not vortex.
4. Incubate at 29-30C for minimum of 2 hours, ideally longer - 4h.
5. Analyse GFP fluorescence immediately. The fluorescence of GFP can be analysed in the reaction mix, without purification. Use Nanodrop, or small volume cuvette fluorimeter.
Scale appropriately if needed.
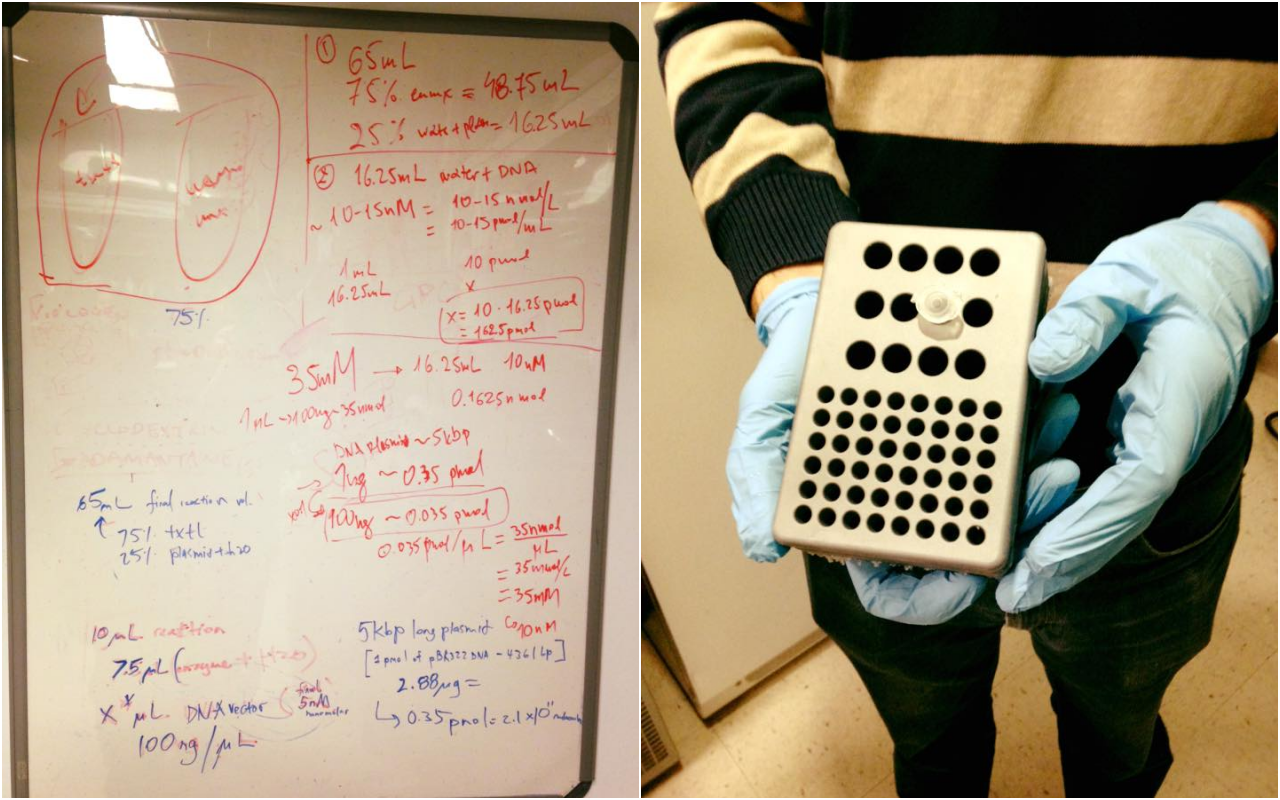
With total 65ul we want 75% enzyme mix and 25% plasmid+water
We know that Kate's GFP plasmid is around 5kbp, so we used this website: https://www.neb.com/tools-and-resources/usage-guidelines/nucleic-acid-data to estimate the pmol of our plasmid. Our plasmid is comparable to pBR322, which is 1ug = 0.35 pmol = 2.1 X 1011 molecules.
After our conclusions we determined that for the 10uL reaction we will use 7.5uL of enzyme mix and 1uL of Kate's GFP plasmid.


We made the mix and then we waiting for about 4 hours.
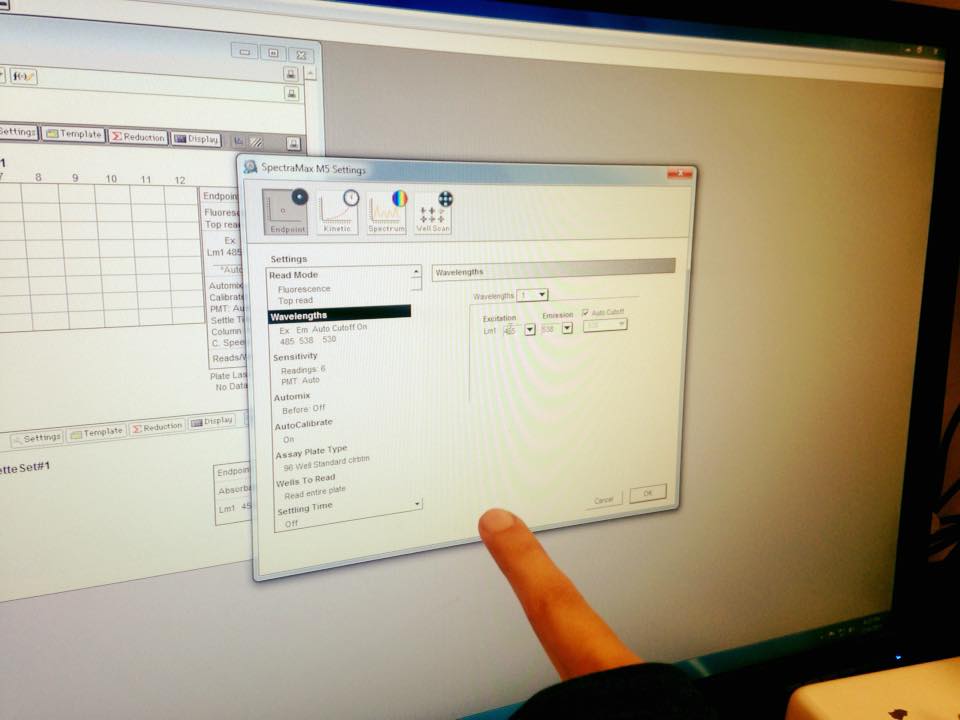
Then we used a spectrophotometer to measure the GFP expression. We compared to a control (well with nothing inside), and there was a noticeable peak about 5 times the control.
We tried to visualize the GFP expression with blue light but were unable to see anything.

/\/\/\/\/\/\/\/\/\/\/\/\/\/\/\/\/\/\/\/\/\/\/\/\/\/\/\/\/\/\/\/\/\/\/\/\/\/\/\/\/\/\/\/
Design-based homework assignment:
Design synthetic cell biosensor, reactor or actuator.
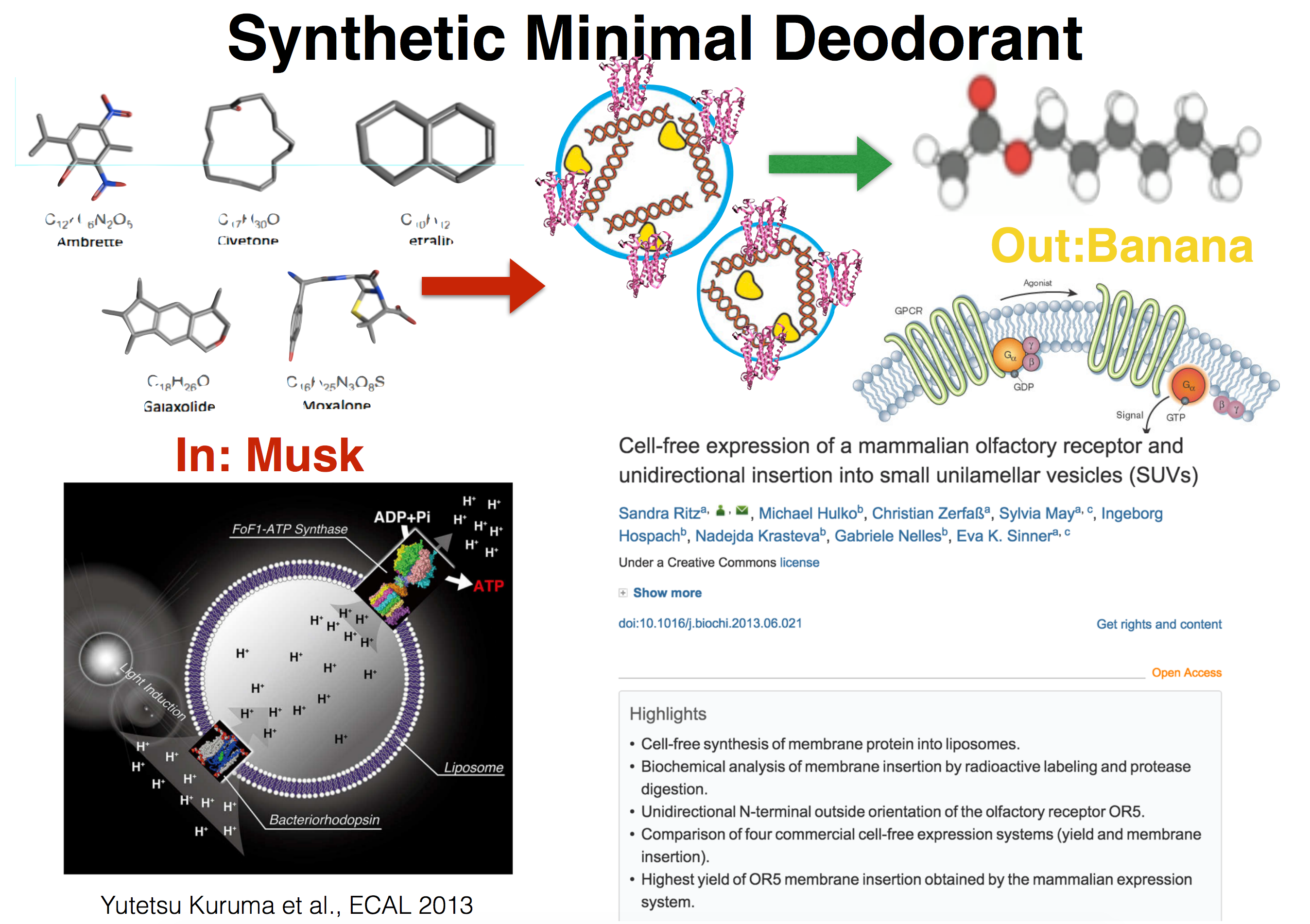
1. Pick a function.
1A What would your synthetic cell do? What is the input and what is the output.
1B Could this function be realized by cell free Tx/Tl alone, without encapsulation?
1C Could this function be realized by genetically modified natural cell?
1D Describe the desired outcome of your synthetic cell operation.
2. Design all components that would need to be part of your synthetic cell.
2A What would be the membrane made of?
2B What would you encapsulate inside? Enzymes, small molecules.
2C Which organism your tx/tl system will come from? is bactrerial OK, or do you need mammalian system for some reason? (hint: for example, if you want to use small molecule modulated promotors, like Tet-ON, you need mammalian).
2D How will your synthetic cell communicate with the environment? (hints: are substrates permeable? or do you need to express membrane channel?)
3. Experimental details
3A List all lipids and genes (bonus: find the specific genes; for example, instead of just saying “small molecule membrane channel” pick actual gene)
3B How will you measure the function of your system?
Bonus: “A picture is worth a thousand words”. The entire homework could be done in one, detailed and annotated scheme showing all the components of your synthetic cell in desired operation.
If possible, please include references (not only original papers, websites are OK too).