DESIGN >> BUILD >> TEST
GROUP HOMEWORK PDF



ASSIGNMENT OVERVIEW
1. What chassis and pathway will you use?
2. How will you measure product formation?
3. What chassis enzymes would you modify?
The product we chose is estrogen (estradiol) beginning with an endogenous cholesterol precursor. It is actually possible to use the cholesterol from egg yolks after some purification. https://pubs.acs.org/doi/abs/10.1021/ed101188q?journalCode=jceda8

First, we researched metabolic pathways for estrogen biosynthesis and the associated enzymes.
https://en.wikipedia.org/wiki/Estrogen#/media/File:Steroidogenesis.svg
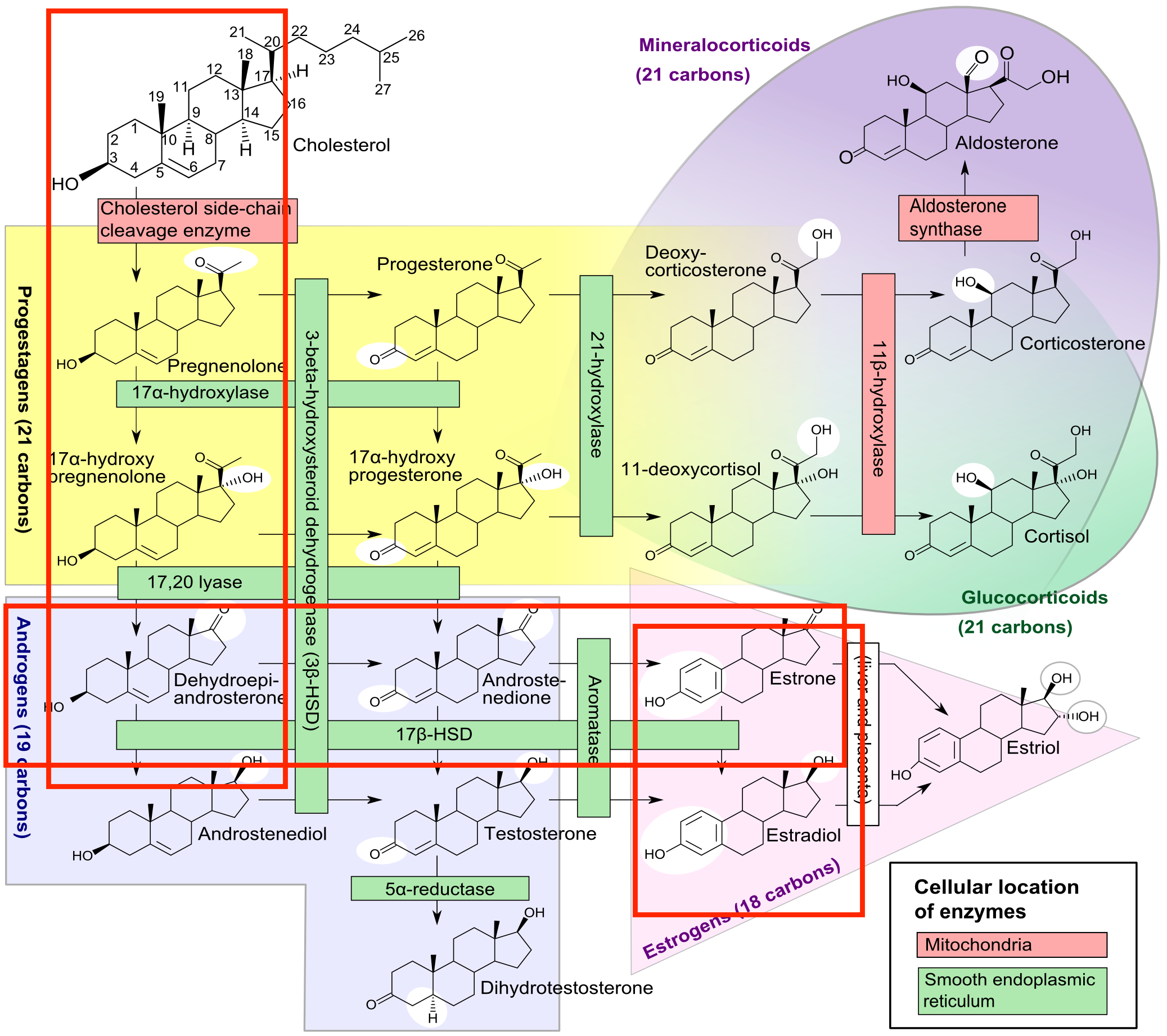
From the pathway above, we can isolate the pathway we want from the below image taken from KEGG. https://www.kegg.jp/kegg-bin/show_pathway?ko00140+K07434
Our desired pathway is highlighted in yellow.
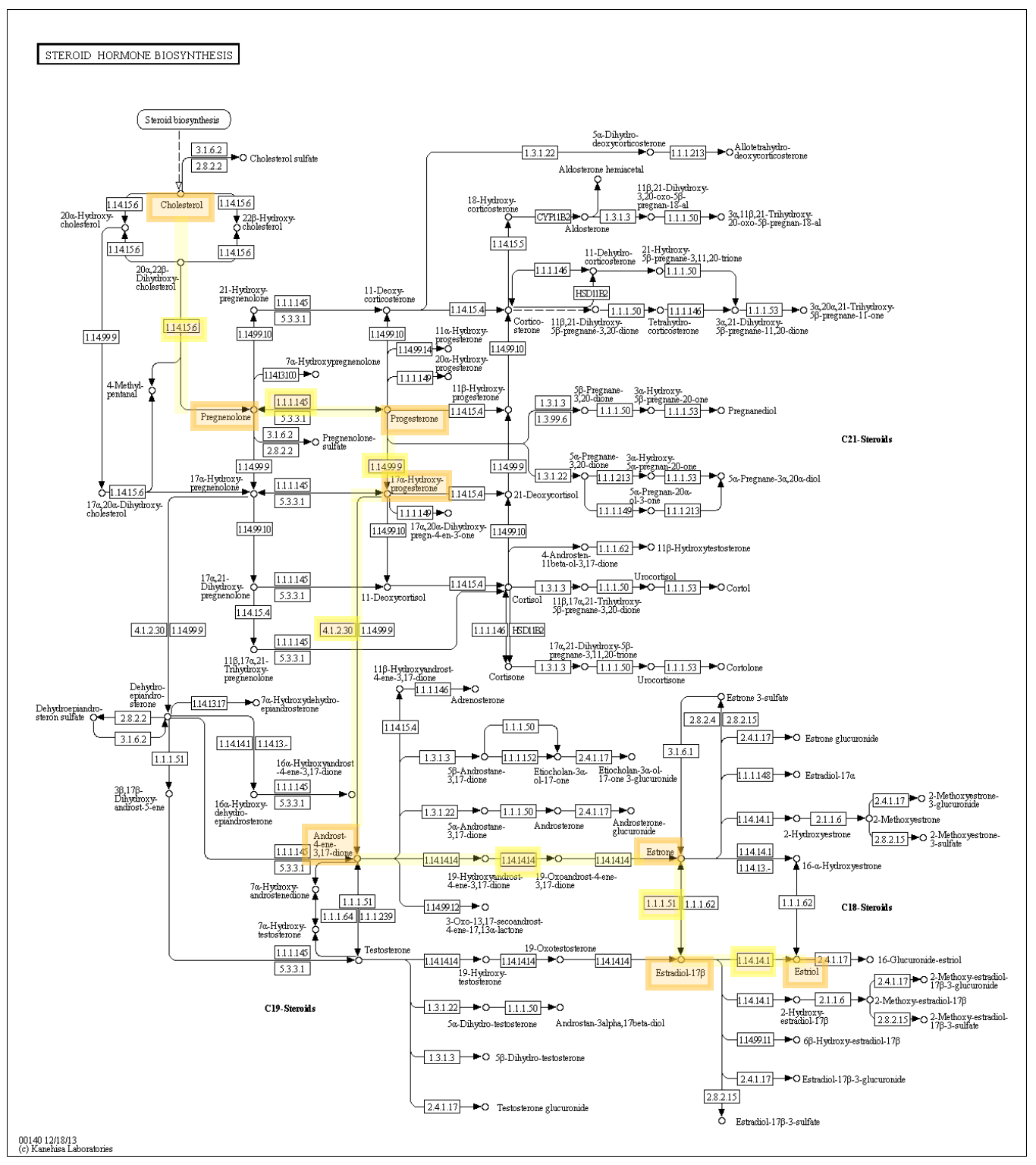
Enzymes (metabolic pathway)
https://www.kegg.jp/dbget-bin/www_bget?ec:1.14.15.6 (cholesterol monooxygenase side-chain-cleaving)
https://www.kegg.jp/dbget-bin/www_bget?ec:1.14.99.9 (CYP17A1)
https://www.kegg.jp/dbget-bin/www_bget?ec:4.1.2.30 (CYP17A1)
https://www.kegg.jp/dbget-bin/www_bget?ec:1.1.1.145 (3beta-hydroxy-Delta5-steroid dehydrogenase)
https://www.kegg.jp/dbget-bin/www_bget?ec:5.3.3.1 (steroid Delta-isomerase)
https://www.kegg.jp/dbget-bin/www_bget?ec:1.14.14.14 (aromatase; CYP19A1)
https://www.kegg.jp/dbget-bin/www_bget?ec:1.1.1.62 (estradiol dehydrogenase)
2. How will you measure product formation?
We can add a GFP reporter protein gene gated by an estrogen receptor.
“Tightly Regulated, betaestradiol DoseDependent Expression System for Yeast” (2000) https://www.biotechniques.com/multimedia/archive/00010/00296st02_10414a.pdf

Other methods are commercially available estrogen tests: