GROUP HOMEWORK PDF


Design Homework: Primer design to linearize plasmid backbone
Software needed for the homework assignment: NuPack
Theory: Use NuPack to help select 18 bp priming sites that amplify a ~2.25 kb region of pUC19 (NEB) immediately upstream of the plac promoter and downstream of the start of lacZalpha. The resulting amplicon excludes the plac promoter and n-terminus of lacZalpha, which enables you to swap in a gene of interest under the control of a promoter of interest using Gibson Assembly later on in your workflow. Design one pair of oligos that prime optimally and another that prime poorly. Describe the PCR thermocycling program that uses Phusion polymerase (NEB) for these pairs. Crucially, determine annealing temperatures and extension times.
Helpful readings:
PCR intro
Primer Design
Nupack task check list:
__Number of strand species >= 2 for combinations of primers and ~50bp regions from pUC19 containing their reverse compliment
__Sizes of complexes >= 2 to check self-hybridization, hybrid primer dimers and annealing to target
__Primer and DNA concentrations are in a range suitable for PCR rx mixture:
__Melt range includes several points in Phusion's annealing temperature range

PRIMER 1:
Scanned 18bp segments, at 4nt step resolution for 5 strand species.
FWD 1: GCCTGGGGTGCCTAATGA
FWD 2: GGGGTGCCTAATGAGTGA
FWD 3: TGCCTAATGAGTGAGCTA
FWD 4: TAATGAGTGAGCTAACTC
FWD 5: GAGTGAGCTAACTCACAT
Specified max complex size at 2 strands (for possible dimers).
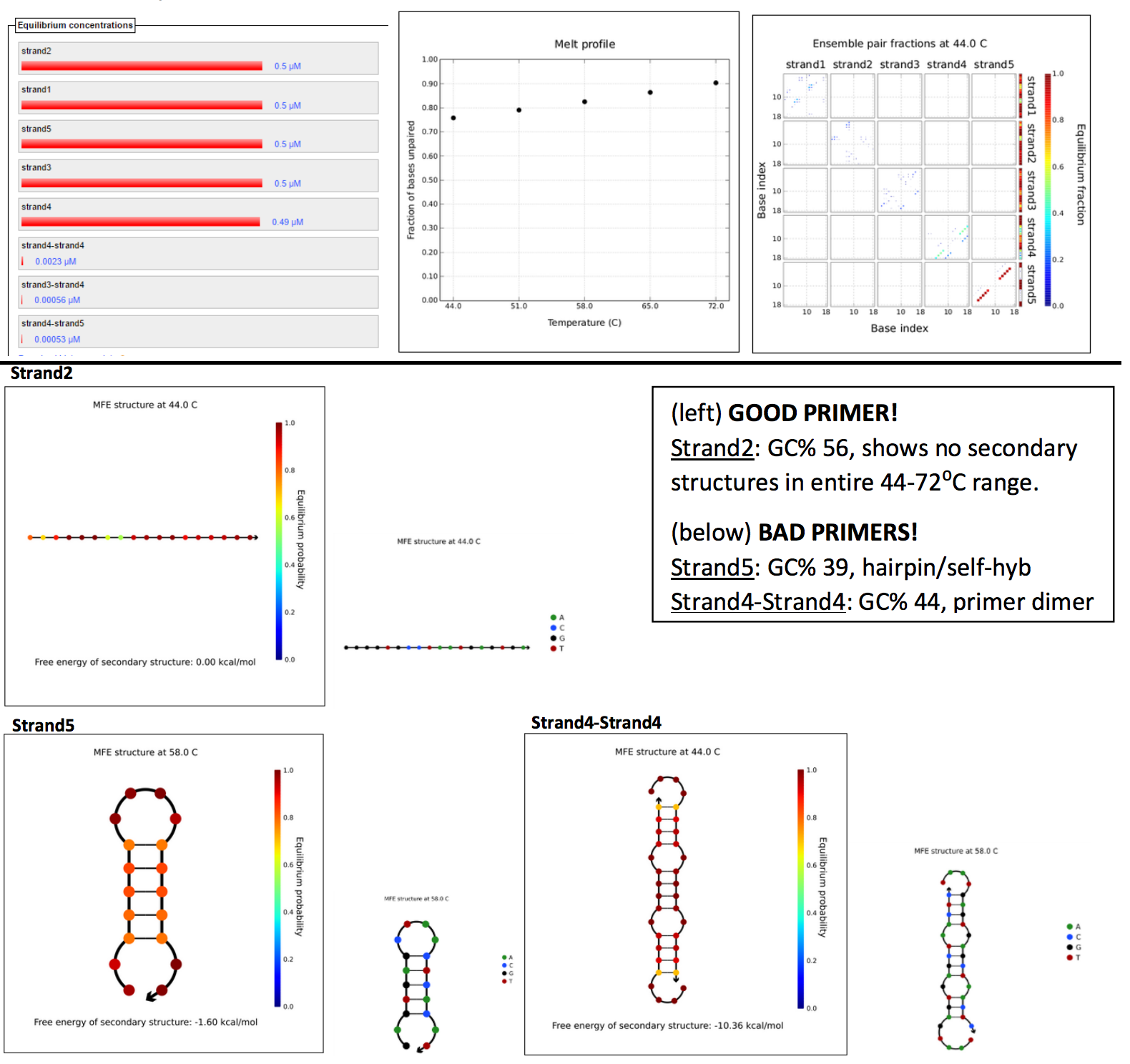
Parameters set by Phusion polymerase, at primer concentration of 0.5µM and several annealing temps ranging between 45-72⁰C (5 temps, with 7⁰C increments), assuming TA is 3-5⁰C below TM. Results: (left to right) Equilibruium concentrations histogram, Melt profile (fraction of bases unpaired), and Ensemble pair fractions.
PRIMER 2:
Scanned 18bp segments, 4nt step res for 5 strand species. (Reverse complement to original sequence.)
REV 1: ATGACCATGATTACGCCA
REV 2: CCATGATTACGCCAAGCT
REV 3: GATTACGCCAAGCTTGCA
REV 4: ACGCCAAGCTTGCATGCC
REV 5: CAAGCTTGCATGCCTGCA
Specified max complex size at 2 strands (for possible dimers).
Parameters set by Phusion polymerase, at primer concentration of 0.5µM and several annealing temps ranging between 45-72⁰C (5 temps, with 7⁰C increments), assuming TA is 3-5⁰C below TM. Results: (left to right) Equilibruium concentrations histogram, Melt profile, and Ensemble pair fractions.

GOOD PRIMER SET:
FWD 2: GGGGTGCCTAATGAGTGA
REV 2: CCATGATTACGCCAAGCT
Similar GC%, and melting temps. No instances of self-annealing or primer dimers between them. Produces a 2.6Kbp amplicon (a little on the large side, target 2.3Kbp)
PCR @ Annealing Temp: 55⁰C
PCR @Extension Time (@ Phusion 15 sec/kb recommendation): 40 sec

BAD PRIMER SET:
FWD 5: GAGTGAGCTAACTCACAT
REV 4: ACGCCAAGCTTGCATGCC
Both primers have GC% out of the 40-60% range, and respectively too low and too high. Because we have to choose a single annealing temperature to use in PCR cycles, the wide range between their GC%/temps adds to the inefficacy of these primers.