GROUP HOMEWORK PDF
EXTRA CREDIT HW PDF
Reading useful for the homework assignment
https://www.nature.com/nature/journal/v440/n7082/abs/nature04586.html
https://www.nature.com/nature/journal/v459/n7245/full/nature08016.html
https://www.sciencemag.org/content/325/5941/725
https://nar.oxfordjournals.org/content/37/15/5001
Software needed for the homework assignment
https://cadnano.org/legacy
caDNAno software tutorials useful for the homework assignment
https://www.youtube.com/watch?v=cwj-4Wj6PMc&feature=youtu.be
https://www.youtube.com/watch?v=EabqNaYAI7o&feature=youtu.be
/\/\/\/\/\/\/\/\/\/\/\/\/\/\/\/\/\/\/\/\/\/\/\/\/\/\/\/\/\/\/\/
DESIGN HOMEWORK:
1. Build flattened Rothemund rectangle in cadDNAno
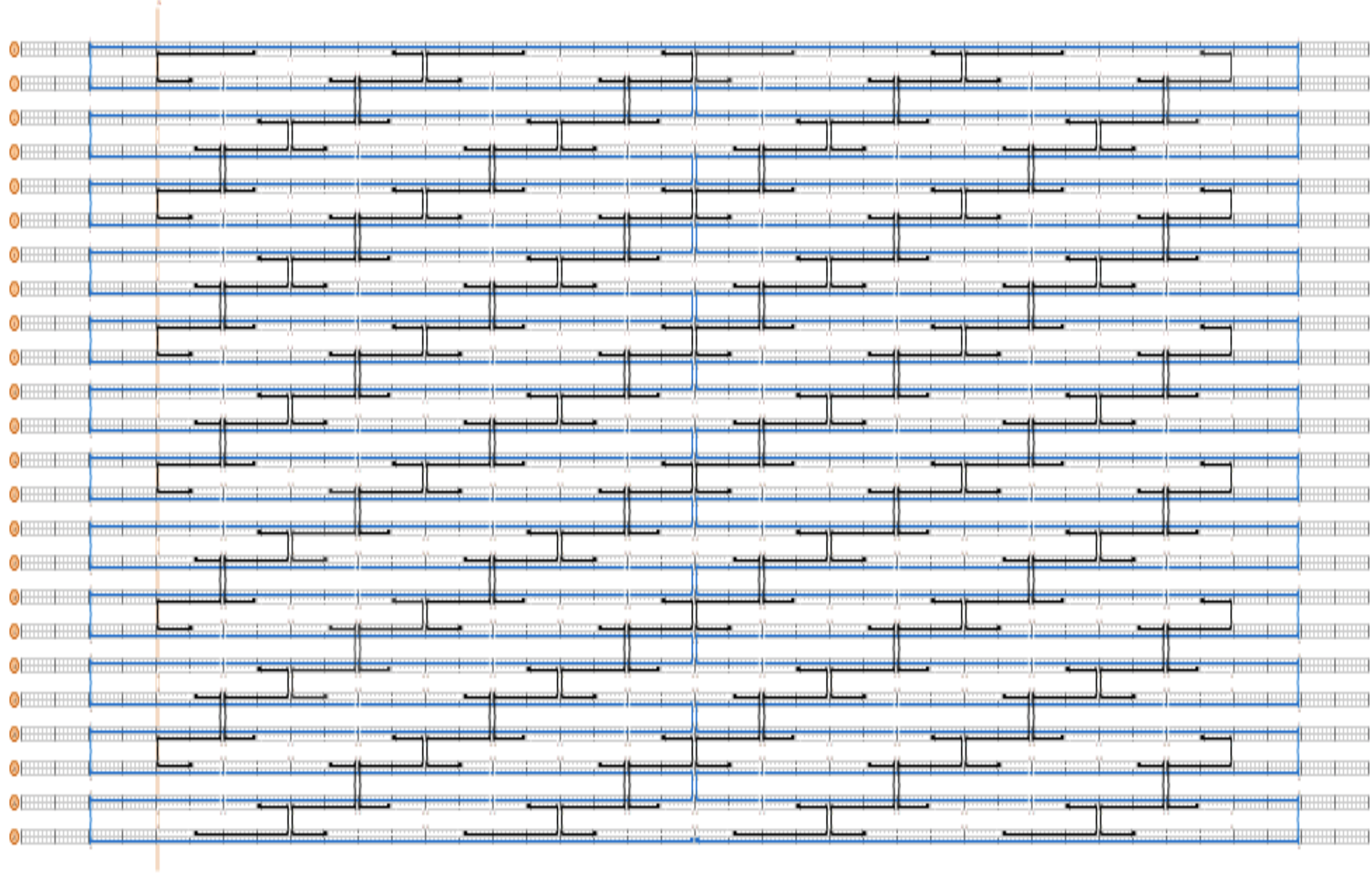
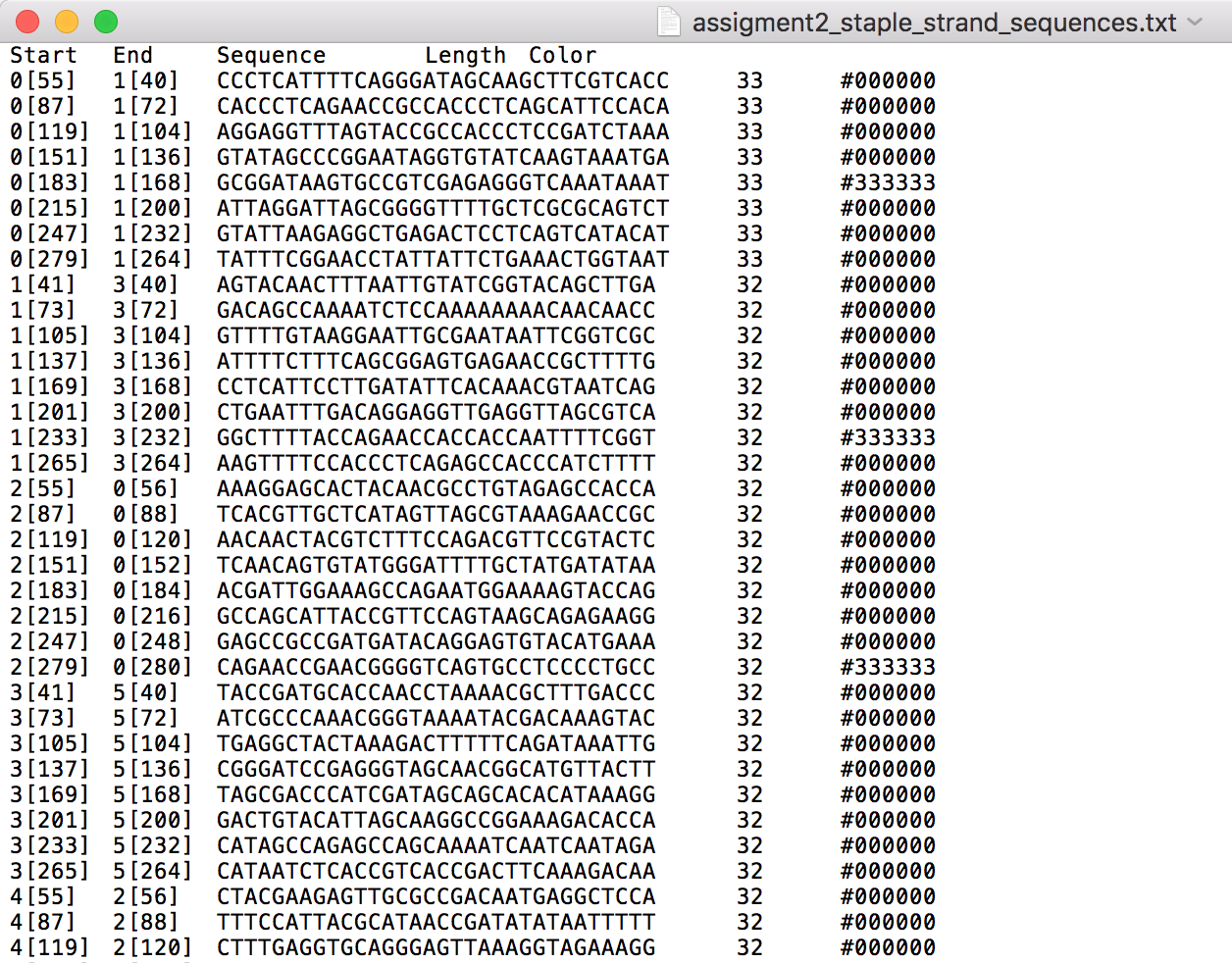
2. Generate staple strand sequences in caDNAno
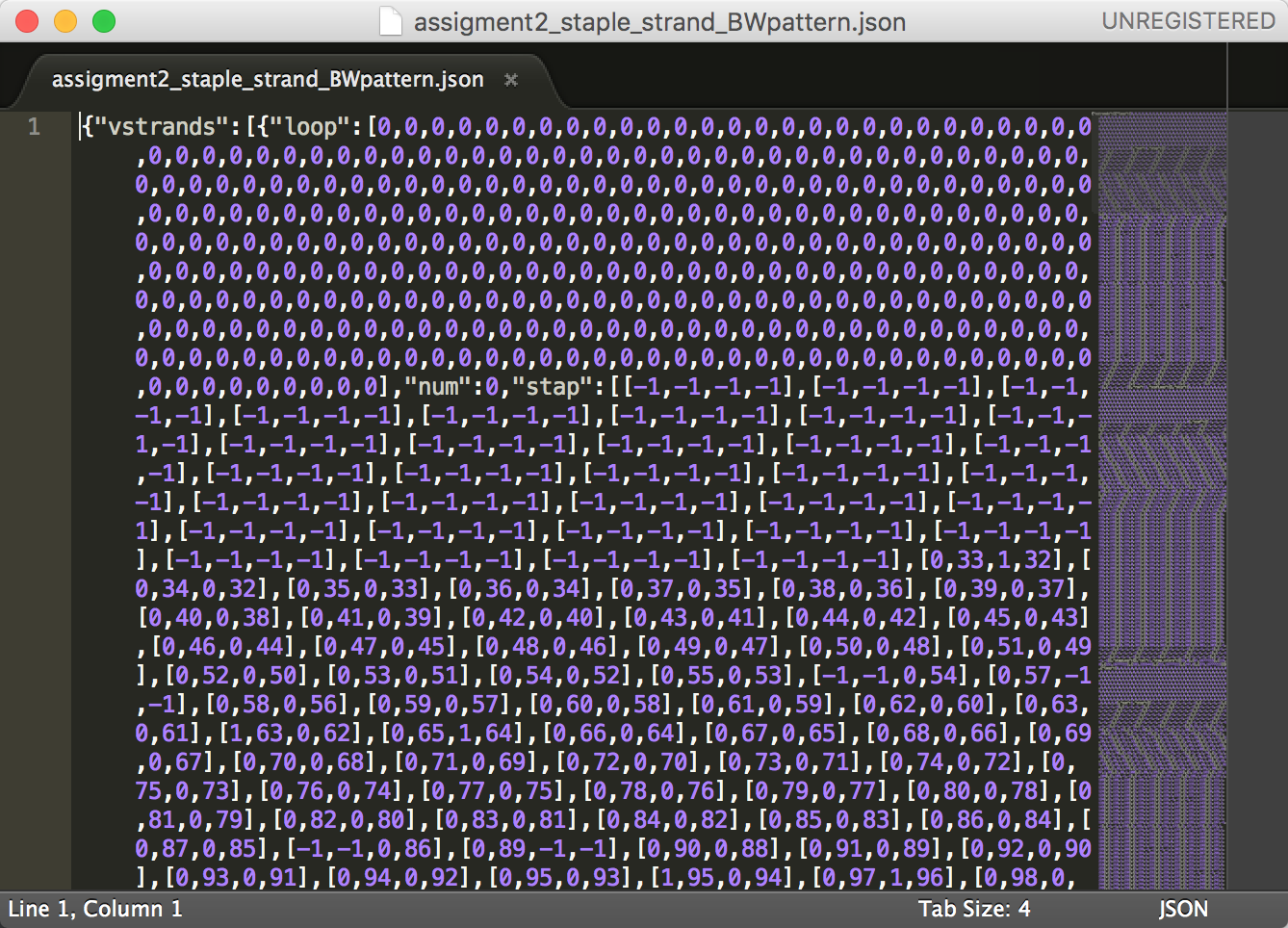
3. Color staple strands to create a black-and-white pattern in caDNAno (below is the JSON file output for the black-and-white pattern)
4. Generate ASCII file black-and-white image

5. Generate list of wells to pipet to generate black-and-white pattern (By parsing the black-and-white ASCII pattern, we selected the appropriate empty and dumbbell staple DNA oligos from the respective 96-well plates)

/\/\/\/\/\/\/\/\/\/\/\/\/\/\/\/\/\/\/\/\/\/\/\/\/\/\/\/\/\/\/\/
EXPERIMENTAL HOMEWORK
__Order from IDT the four 96-well plates needed to render 1-bit patterns:: ~$1500–$2000 for this set of strands on the 10 nmol scale (request 6 nmol total: 60 µL at 100 µM in water). Assuming each pattern will consume 10 pmol per strand, this will be enough material to fold 600 patterns.
__Pipet strands (10 pmol each) to generate all patterns submitted by teams in the class
__Fold patterns in a thermocycler in a volume of 100 µL per pattern
__Check to see that the structures fold correctly by agarose-gel analysis of 15 µL aliquot
__Find a partner (commercial or otherwise) willing to do AFM imaging of your samples
__Submit your samples to that partner (only 1 µL required for imaging)